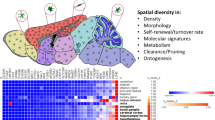
Abstract
Microglia have critical roles in neural development, homeostasis and neuroinflammation and are increasingly implicated in age-related neurological dysfunction. Neurodegeneration often occurs in disease-specific, spatially restricted patterns, the origins of which are unknown. We performed to our knowledge the first genome-wide analysis of microglia from discrete brain regions across the adult lifespan of the mouse, and found that microglia have distinct region-dependent transcriptional identities and age in a regionally variable manner. In the young adult brain, differences in bioenergetic and immunoregulatory pathways were the major sources of heterogeneity and suggested that cerebellar and hippocampal microglia exist in a more immune-vigilant state. Immune function correlated with regional transcriptional patterns. Augmentation of the distinct cerebellar immunophenotype and a contrasting loss in distinction of the hippocampal phenotype among forebrain regions were key features during aging. Microglial diversity may enable regionally localized homeostatic functions but could also underlie region-specific sensitivities to microglial dysregulation and involvement in age-related neurodegeneration.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Prinz, M. & Mildner, A. Microglia in the CNS: immigrants from another world. Glia 59, 177–187 (2011).
Prinz, M. & Priller, J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat. Rev. Neurosci. 15, 300–312 (2014).
Paolicelli, R.C. et al. Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011).
Schafer, D.P. et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 (2012).
Coull, J.A.M. et al. BDNF from microglia causes the shift in neuronal anion gradient underlying neuropathic pain. Nature 438, 1017–1021 (2005).
Neumann, H., Kotter, M.R. & Franklin, R.J.M. Debris clearance by microglia: an essential link between degeneration and regeneration. Brain 132, 288–295 (2009).
Lalancette-Hébert, M., Gowing, G., Simard, A., Weng, Y.C. & Kriz, J. Selective ablation of proliferating microglial cells exacerbates ischemic injury in the brain. J. Neurosci. 27, 2596–2605 (2007).
Fenn, A.M., Hall, J.C.E., Gensel, J.C., Popovich, P.G. & Godbout, J.P. IL-4 signaling drives a unique arginase+/IL-1β+ microglia phenotype and recruits macrophages to the inflammatory CNS: consequences of age-related deficits in IL-4Rα after traumatic spinal cord injury. J. Neurosci. 34, 8904–8917 (2014).
Cardona, A.E. et al. Control of microglial neurotoxicity by the fractalkine receptor. Nat. Neurosci. 9, 917–924 (2006).
Biber, K., Neumann, H., Inoue, K. & Boddeke, H.W. Neuronal 'On' and 'Off' signals control microglia. Trends Neurosci. 30, 596–602 (2007).
Hickman, S.E. et al. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16, 1896–1905 (2013).
Butovsky, O. et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014).
Lawson, L.J., Perry, V.H., Dri, P. & Gordon, S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 39, 151–170 (1990).
de Haas, A.H. Boddeke, H.W.G.M. & Biber, K. Region-specific expression of immunoregulatory proteins on microglia in the healthy CNS. Glia 56, 888–894 (2008).
Wang, Y. et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 13, 753–760 (2012).
Greter, M. et al. Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity 37, 1050–1060 (2012).
Karch, C.M. & Goate, A.M. Alzheimer's disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry 77, 43−51 (2015).
Yao, P. et al. Coexpression networks identify brain region-specific enhancer RNAs in the human brain. Nat. Neurosci. 18, 1168–1174 (2015).
Chiu, I.M. et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Reports 4, 385–401 (2013).
Freeman, T.C. et al. Construction, visualisation, and clustering of transcription networks from microarray expression data. PLoS Comput. Biol. 3, 2032–2042 (2007).
Hume, D.A., Summers, K.M., Raza, S., Baillie, J.K. & Freeman, T.C. Functional clustering and lineage markers: insights into cellular differentiation and gene function from large-scale microarray studies of purified primary cell populations. Genomics 95, 328–338 (2010).
Takaoka, A. et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 (2007).
Ghesquière, B., Wong, B.W., Kuchnio, A. & Carmeliet, P. Metabolism of stromal and immune cells in health and disease. Nature 511, 167–176 (2014).
Linnartz, B., Wang, Y. & Neumann, H. Microglial immunoreceptor tyrosine-based activation and inhibition motif signaling in neuroinflammation. Int. J. Alzheimers Dis. 2010, 587463 (2010).
Freilich, R.W., Woodbury, M.E. & Ikezu, T. Integrated expression profiles of mRNA and miRNA in polarized primary murine microglia. PLoS One 8, e79416 (2013).
Mathelier, A. et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 42, D142–D147 (2014).
Frith, M.C. et al. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 32, 1372–1381 (2004).
Huss, J.M., Torra, I.P., Staels, B., Giguère, V. & Kelly, D.P. Estrogen-related receptor α directs peroxisome proliferator-activated receptor α signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 24, 9079–9091 (2004).
Mootha, V.K. et al. Erralpha and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA 101, 6570–6575 (2004).
Pearen, M.A. & Muscat, G.E.O. Minireview: Nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol. Endocrinol. 24, 1891–1903 (2010).
Archer, M.C. Role of sp transcription factors in the regulation of cancer cell metabolism. Genes Cancer 2, 712–719 (2011).
Francis, G.A., Fayard, E., Picard, F. & Auwerx, J. Nuclear receptors and the control of metabolism. Annu. Rev. Physiol. 65, 261–311 (2003).
Griffin, M.J. et al. Early B-cell factor-1 (EBF1) is a key regulator of metabolic and inflammatory signaling pathways in mature adipocytes. J. Biol. Chem. 288, 35925–35939 (2013).
Chen, B.-S., Yang, S.-K., Lan, C.-Y. & Chuang, Y.-J. A systems biology approach to construct the gene regulatory network of systemic inflammation via microarray and databases mining. BMC Med. Genomics 1, 46 (2008).
Natoli, G., Ghisletti, S. & Barozzi, I. The genomic landscapes of inflammation. Genes Dev. 25, 101–106 (2011).
Schnell, L., Fearn, S., Klassen, H., Schwab, M.E. & Perry, V.H. Acute inflammatory responses to mechanical lesions in the CNS: differences between brain and spinal cord. Eur. J. Neurosci. 11, 3648–3658 (1999).
Hart, A.D., Wyttenbach, A., Perry, V.H. & Teeling, J.L. Age related changes in microglial phenotype vary between CNS regions: grey versus white matter differences. Brain Behav. Immun. 26, 754–765 (2012).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
Bernhart, E. et al. Lysophosphatidic acid receptor activation affects the C13NJ microglia cell line proteome leading to alterations in glycolysis, motility, and cytoskeletal architecture. Proteomics 10, 141–158 (2010).
Stocking, C. & Kozak, C.A. Murine endogenous retroviruses. Cell. Mol. Life Sci. 65, 3383–3398 (2008).
Yu, P. et al. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity 37, 867–879 (2012).
Lee, K.H., Horiuchi, M., Itoh, T., Greenhalgh, D.G. & Cho, K. Cerebellum-specific and age-dependent expression of an endogenous retrovirus with intact coding potential. Retrovirology 8, 82 (2011).
Baillie, J.K. et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479, 534–537 (2011).
Johnson-Wood, K. et al. Amyloid precursor protein processing and A beta42 deposition in a transgenic mouse model of Alzheimer disease. Proc. Natl. Acad. Sci. USA 94, 1550–1555 (1997).
Damani, M.R. et al. Age-related alterations in the dynamic behavior of microglia. Aging Cell 10, 263–276 (2011).
Streit, W.J., Braak, H., Xue, Q.-S. & Bechmann, I. Dystrophic (senescent) rather than activated microglial cells are associated with tau pathology and likely precede neurodegeneration in Alzheimer's disease. Acta Neuropathol. 118, 475–485 (2009).
Sleimen-Malkoun, R., Temprado, J.J. & Hong, S.L. Aging induced loss of complexity and dedifferentiation: consequences for coordination dynamics within and between brain, muscular and behavioral levels. Front. Aging Neurosci. 6, 140 (2014).
Goh, J.O.S. Functional dedifferentiation and altered connectivity in older adults: neural accounts of cognitive aging. Aging Dis. 2, 30–48 (2011).
Park, D.C. et al. Aging reduces neural specialization in ventral visual cortex. Proc. Natl. Acad. Sci. USA 101, 13091–13095 (2004).
Davies, L.C., Jenkins, S.J., Allen, J.E. & Taylor, P.R. Tissue-resident macrophages. Nat. Immunol. 14, 986–995 (2013).
Lein, E.S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Huang, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5, e13984 (2010).
Cline, M.S. et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 (2007).
Eden, E., Navon, R., Steinfeld, I., Lipson, D. & Yakhini, Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009).
Acknowledgements
We thank members of Edinburgh Genomics, The University of Edinburgh, for performing microarray assays, B. Fleming for technical assistance with microglial purification and Y.-T. Lai for help optimizing regional brain dissection. This work was funded by a PhD scholarship from the Darwin Trust of Edinburgh to K Grabert and grants from the Biotechnology and Biological Sciences Research Council (BBSRC; BB/J004332/1) and the Medical Research Council (MRC; MR/L003384/1). The Roslin Institute and Edinburgh Genomics are partly supported through core grants from the Natural Environment Research Council (R8/H10/56), MRC (MR/K001744/1) and BBSRC (BB/J004243/1, BB/J004332/1).
Author information
Authors and Affiliations
Contributions
B.W.M. and K.M.S. conceived the study; B.W.M., K.M.S., K.G. and T.C.F. designed experiments; K.G., B.W.M. performed experiments and analyzed data; T.M. advised on and contributed to data visualization; M.H.K. and M.P.S. advised on and contributed to bacterial assay; S.C. and J.K.B. advised on and performed analysis of transcriptional regulators; B.W.M., K.M.S. and K.G. wrote the paper; all authors contributed to data interpretation and editing of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Validation of regional microglial isolation
(a) Workflow showing key steps in multi-region microglial isolation. (b) Brain cell suspensions were prepared from Csf1r-EGFP reporter mice and the expression of CD11b on GFP+ cells (i.e. microglia) assessed by flow cytometry. Complete overlap between CD11b and GFP is evident. Data are representative of three independent cell preparations (c) CD11b and F4/80 expression were assessed by flow cytometry on the selected and non-selected cell fractions after immunomagnetic separation. Single populations of CD11b+ and F4/80+ cells in the selected fraction and the absence of staining in the non-selected fraction indicates the specificity and efficiency of extraction. Data are representative of four independent samples, each from tissue prepared from eight mice (d) Microarray expression profiles of selected genes in mixed brain cell homogenates prepared from distinct brain regions. Genes chosen have previously established regional patterns of expression in neuronal subpopulations. The expression profiles in brain homogenates showed the expected regional pattern as validated by correlation with spatial expression profiles in the Allen Brain Atlas (http://mouse.brain-map.org/). URLs for images are provided in the Methods section. Image credit: Allen Institute for Brain Science. Data show mean ± SEM, n = 2 independent samples (mice). Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum.
Supplementary Figure 2 Network analysis of regional microglial gene expression
Network analysis of regional microglial gene expression. (a) A transcript-to-transcript correlation network graph of transcripts significantly differentially expressed by brain region was generated in BioLayout Express3D. Nodes represent transcripts (probesets) and edges the degree of correlation between them. The network graph was clustered using a Markov clustering algorithm. Key network graph metrics and description of expression pattern for clusters determined with Pearson correlation threshold of 0.80 or 0.90 are shown. (b) Network diagram showing similar overall topography with Pearson correlation threshold of 0.80 or 0.90.
Supplementary Figure 3 Networks centered on interferon pathways are enriched in cluster 3
Pathway analysis in Ingenuity identified an enriched IRF7-centred interferon network within cluster 3 from the network graph of differentially expressed transcripts generated in BioLayout Express3D. Filled circle/oval: genes represented in the cluster 3 dataset; empty circle/oval: genes not contained in cluster 3 but associated with the network; dashed arrow: indirect connection, solid arrow: direct connection; multiple circles: complex.
Supplementary Figure 4 Enrichment of an antigen presentation pathway in cluster 3
KEGG pathway analysis using DAVID identified antigen processing and presentation as an enriched pathway within cluster 3 from the network graph of differentially expressed transcripts generated in BioLayout Express3D. Stars denote pathway genes present in cluster 3.
Supplementary Figure 5 Regional heterogeneity in expression of microglial cell surface genes
(a) Transcripts differentially expressed according to brain region were analysed for enrichment (p < 0.05 with Benjamini correction) of Gene Ontology (GO) Cellular Component and visualised in GOrilla. (b) Heat map showing the expression pattern of microglial “sensome” genes that were differentially expressed according to brain region in our dataset. Row Z-score intensities represent the mean of four independent samples per region with red indicating high probeset expression and blue low expression. (c) Microarray expression levels of genes encoding cell surface proteins transducing microglial “off” signals. Data show mean ± SD, n = 4 independent samples, each pooled from tissue from eight mice. *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA with Bonferroni correction. Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum. Specific p values for all statistical comparisons are presented in Supplementary Table 13.
Supplementary Figure 6 Regional variations in age-related depression of young adult microglial signature
(a) Fold-changes in expression of selected microglial signature genes between 4 and 22 months in each brain region. Data show mean ± SEM, n = 4 independent samples, each pooled from tissue from eight mice. *p < 0.05, **p < 0.01, ***p < 0.001 vs Cbm, one-way ANOVA with Dunnett post-test. (b-d) Expression profiles of (b) Tgfbr family genes, (c) systemic macrophage signature genes with Itgam as reference, and (d) genes upregulated on activated microglia/macrophages during ageing in each brain region. Data show mean ± SEM, n = 4 independent samples, each pooled from tissue from eight mice. *p < 0.05, **p < 0.01, ***p < 0.001, two-way ANOVA with Bonferroni post-test. Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum. Specific p values for all statistical comparisons are presented in Supplementary Table 13.
Supplementary Figure 7 Regional expression pattern of Mela in mixed brain cell homogenates at 4 months of age
Expression was determined from microarray analysis of mixed brain cell homogenates dissected from discrete brain regions. Data show mean ± SEM, n = 2 independent samples (mice). *p < 0.05 vs all other regions, one-way ANOVA with Bonferroni correction. Str, striatum; Hpp, hippocampus; Ctx, cerebral cortex; Cbm, cerebellum. Specific p values for all statistical comparisons are presented in Supplementary Table 13.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 1232 kb)
Supplementary Methods Checklist
(PDF 451 kb)
Supplementary Table 1
Probesets differentially regulated by brain region (XLSX 195 kb)
Supplementary Table 2
Over-represented GO biological processes at 4 months (XLSX 14 kb)
Supplementary Table 3
Annotation of Enrichment Map nodes at 4 months (XLSX 13 kb)
Supplementary Table 4
Transcript composition of clusters of gene co-expression at 4 months (XLSX 78 kb)
Supplementary Table 5
Over-represented GO biological processes in immune gene co-expression cluster at 4 months (XLSX 14 kb)
Supplementary Table 6
Ingenuity analysis of upstream regulators in immune gene co-expression cluster at 4 months (XLSX 11 kb)
Supplementary Table 7
Over-represented KEGG pathways in immune gene co-expression cluster at 4 months (XLSX 12 kb)
Supplementary Table 8
Over-represented GO biological processes in bioenergetics gene co-expression cluster 3 at 4 months (XLSX 14 kb)
Supplementary Table 9
Transcriptional regulators of region-dependent co-expression networks at 4 months (XLSX 11 kb)
Supplementary Table 10
Over-represented GO biological processes in regionally differentially regulated sensome genes at 4 months (XLSX 13 kb)
Supplementary Table 11
Over-represented GO biological processes in gene co-expression cluster 2 from ageing datasets (XLSX 11 kb)
Supplementary Table 12
Over-represented GO biological processes in age down-regulated genes from hippocampal microglia (XLSX 10 kb)
Supplementary Table 13
Tables of specific p values for all statistical comparisons (XLSX 54 kb)
Rights and permissions
About this article
Cite this article
Grabert, K., Michoel, T., Karavolos, M. et al. Microglial brain region−dependent diversity and selective regional sensitivities to aging. Nat Neurosci 19, 504–516 (2016). https://doi.org/10.1038/nn.4222
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4222
This article is cited by
-
Tmem119 expression is downregulated in a subset of brain metastasis-associated microglia
BMC Neuroscience (2024)
-
Susceptibility to acute cognitive dysfunction in aged mice is underpinned by reduced white matter integrity and microgliosis
Communications Biology (2024)
-
Microglia regulation of central nervous system myelin health and regeneration
Nature Reviews Immunology (2024)
-
Recent advances and current challenges of new approach methodologies in developmental and adult neurotoxicity testing
Archives of Toxicology (2024)
-
hESC-derived striatal progenitors grafted into a Huntington’s disease rat model support long-term functional motor recovery by differentiating, self-organizing and connecting into the lesioned striatum
Stem Cell Research & Therapy (2023)